Last Wednesday, I listened to a really cool talk by Dr. Rakesh Bam. He’s working on developing a new method to break through in ultrasound molecular imaging for early breast cancer detection. There are 3 stages of breast cancer that are survivable; the fourth and final stage is really hard to battle against as that is when metastasis occurs. Thus, early detection of the cancer is key.
Mammography, ultrasounds, and MRIs are some of the ways to image breast tissue and detect breast cancer. Mammography is good for women with scattered fibroglandular tissue. It’s limited dealing with very dense breasts. Ultrasound uses 720 kHz sound waves to image soft tissue. However, differentiating signal from noise can be hard sometimes, and there are false positives in this screening. Contrast agents are one way to deal with this problem.
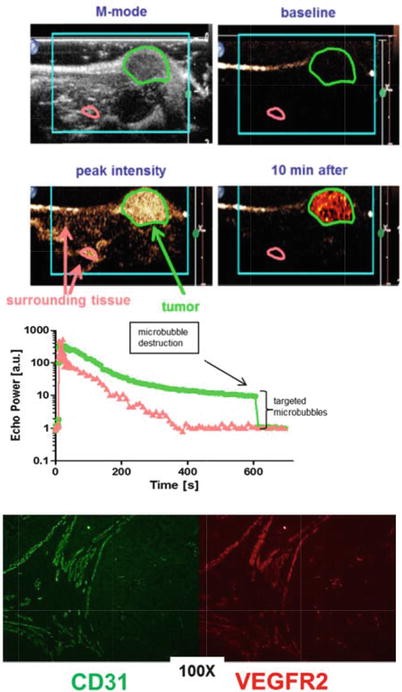
Dr. Bam is experimenting with microbubbles (going down the same avenue as contrast agents) to see if they can be a viable method of breast cancer detection. He uses targeted microbubbles; these microbubbles (gas-filled bubbles that are made of phospholipids) are tagged with receptors that can bind to ligands on cancer cells in the endothelial tissue of blood vessels. CD31, KDR, and B7-H3 are some of the biomarkers that Dr. Bam is working with.
In the tumor microenvironment, there are blood vessels supplying the cancer. The goal is for the microbubbles to bind to a cancer biomarker such as CD31 or KDR or B7-H3 in the growing blood vessels in the location of the tumor. These microbubbles carrying the contrast agent should enable better, more precise visualization of the tumor in the ultrasound. KDR and B7-H3 in the capillaries has been shown to be a good biomarker for tumor visualization in clinical trials. They tend to be overexpressed in cancer cells in blood vessels in the tumor, so there is better signal to noise ratio there (minimal or no binding to normal tissues as they don’t express these biomarkers compared to the cancer cells).
Note: The above image is from https://link.springer.com/chapter/10.1007/978-3-319-42202-2_14.
Dr. Bam has experimented with his microbubbles in murine models with random tumors with good results. He has been able to replace the antibody (serving as the receptor) latched onto the microbubble with a smaller yet still effective protein (antibodies are expensive to produce). This affinity protein also lets the microbubble circulate for minutes as opposed to the 48 hours using antibodies allowing for faster imaging.
In the future, Dr. Bam plans to experiment with other kinds of microbubbles and replace the biotin-streptavidin linkage (which connects the microbubble with the receptor) with a nontoxic alternative.