Today, proteins, miRNA, cancer mutations, and more databases were discussed. X-ray crystallography and nuclear magnetic resonance (NMR) are common methods used to determine protein structure. NCBI, PDB, and COSMIC-3D (druggability and mutation cluster features) are useful protein databases. miRNAs target 3′ UTR and restrict protein expression. They also regulate alternative splicing and mRNA methylation. shRNA is double-stranded and are research tools employed in gene knockouts. The biogenesis of mRNA is depicted in the diagram above. Targetscan and Mirtarbase are miRNA databases where you can view predicted miRNA targets. PhosphoSitePlus, Prosite, PFAM, CDART, Eukaryotic Linear Motif are additional protein databases with information on domains and post-translational modifications. Dep Map and Project Achilles report results on cancer cell proliferation when particular genes are knocked out by CRISPR.
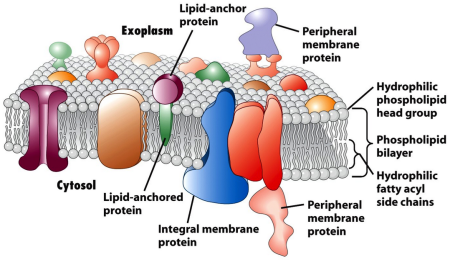
I also presented research on my gene c3orf80, which is a single pass transmembrane protein with glycosylation at amino acid 57. Through analysis of microarrays, protein motifs, conserved domains, tissue expression, predicted transcription factors, knockouts in cancer cell lines, mutation distribution, post-translational modifications, and more, I deduced that it’s an oncogene involved in some kinds of pancreatic cancer, may be involved in cell differentiation (transcription factor STAT4 is involved in T helper cell differentiation along with the related STAT1), possibly could function in Notch pathway due to upregulation of JAG1 and c3orf80 (microarray data), and cervical cancer (HeLa S3 cells can grow in suspension when this gene is upregulated). It’s highest relative tissue expression is in the cerebral cortex in areas like the primary visual center; I’m not sure what it’s exactly doing there. It’s interesting to note that it has a conserved domain from sodium-potassium pumps; however, it can’t function as one as it only goes through the membrane once. Obviously, these are all hypotheses based on research through bioinformatics tools online, and lab work would be needed to confirm or debunk these theories. Nonetheless, bioinformatics is a powerful tool to guide scientists’ exploration in uncharted waters, and I certainly enjoyed the challenge!